STELLA.ai
May 2024 - September 2024
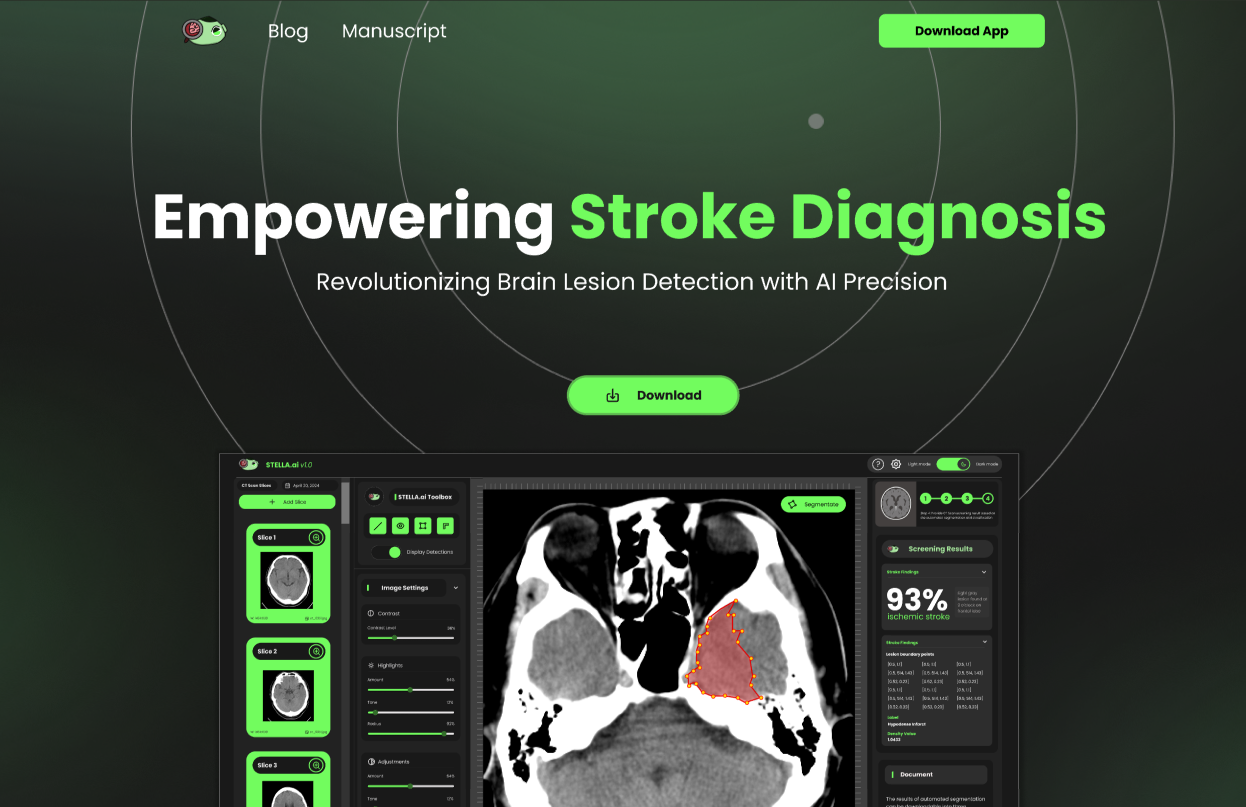
Description
STELLA.ai (Stroke Tomography for Enhanced Lesion Learning Analysis) is an intelligent system developed to automate the detection and segmentation of brain lesions from CT scans. It specializes in identifying ischemic and hemorrhagic strokes. The application integrates artificial intelligence to address challenges in early stroke detection. Its key focus is to provide reliable results that can assist medical professionals. The system is designed to be accurate, efficient, and user-friendly, making it a valuable tool in healthcare diagnostics.
Problem
Detecting strokes accurately and promptly is a significant challenge in medical imaging. Manual analysis of CT scans can take time and is prone to human error, delaying treatment. This delay can be detrimental, as strokes require immediate medical attention to prevent severe damage. Additionally, distinguishing between ischemic and hemorrhagic strokes requires expertise and precise tools. Many existing solutions lack the efficiency needed for real-time applications in clinical environments. Addressing these challenges was the driving force behind STELLA.ai's development.
Technical Details
STELLA.ai utilizes a U-Net Convolutional Neural Network (CNN) to perform precise segmentation of stroke lesions in CT scans. The system preprocesses data using OpenCV and classifies stroke types based on intensity values. A Python-based backend powered by FastAPI ensures smooth integration and real-time responsiveness. It employs TensorFlow for the deep learning model and leverages Electron to deliver a cross-platform desktop application. These technologies work in tandem to ensure accurate results, high performance, and user accessibility. The model's training included extensive preprocessing to handle diverse CT scan qualities and enhance prediction reliability.
My Role
As the lead developer, I spearheaded the design and implementation of the U-Net model for lesion segmentation. My responsibilities included optimizing the preprocessing pipeline using OpenCV and developing the Python backend with FastAPI. I also integrated the stroke classification module based on Hounsfield Unit (HU) ranges. In addition, I designed the desktop application interface using Electron to ensure user-friendly navigation. I worked closely on data augmentation strategies to enhance model training and collaborated with peers to evaluate system performance. My role was pivotal in ensuring the accuracy and reliability of the system.
Challenges
One of the primary challenges was obtaining a diverse dataset that included sufficient ischemic and hemorrhagic stroke samples. Training the model to generalize across CT scans of varying quality was another hurdle. The system needed to be highly accurate while maintaining real-time performance. Another difficulty was creating a robust stroke classification mechanism based on Hounsfield Unit (HU) values. Implementing a cross-platform desktop application that integrates seamlessly with the backend required meticulous planning. These challenges were addressed through extensive research, testing, and iterative development.
Screenshots
Conclusion
STELLA.ai has shown the transformative potential of AI in healthcare, particularly in medical imaging. The system provides a reliable tool for healthcare professionals to detect and classify strokes swiftly and accurately. It enhances diagnostic efficiency, contributing to improved patient outcomes. The project highlights how AI can assist in critical decision-making processes in medical settings. By addressing existing gaps in stroke detection, STELLA.ai sets a foundation for future advancements in AI-driven medical diagnostics. The project successfully demonstrates how technology can improve access to quality healthcare solutions.